
Several advances in PCR have dramatically reduced the duration of PCR amplification reactions. Yellow box indicates optimal temperatures. Arrows indicate the annealing temperature that provided the highest specificity while maintaining good yield. Four different primer sets (A, B, C, and D) were designed and tested for amplification. All reactions were evaluated in a single run. Consult the PCR Assay Troubleshooting page for more help. If satisfactory results are not obtained at any annealing temperature, additional optimization steps must be taken, and primer redesign may be necessary. A sample annealing temperature optimization experiment is shown in Figure 1. The optimal annealing temperature is the one that results in the highest yield with no nonspecific amplification. If nonspecific amplification has occurred, additional bands will appear on the gel. Analyze the results using agarose gel electrophoresis. To find the optimal annealing temperature for your reaction, test a range of temperatures above and below the calculated T m of the primers. The gradient feature allows you to test a range of temperatures simultaneously, optimizing the annealing temperature in a single experiment. All Bio-Rad thermal cyclers offer a gradient feature. The optimal annealing temperature for an assay can be easily determined using PCR instruments that have a thermal gradient feature. Similar time-consuming tests may also be required to optimize the denaturation temperature. This involves repeating a reaction at many different temperatures. Even after calculating the T m of a primer, you may need to determine the annealing temperature empirically. On the other hand, setting the annealing temperature too high may reduce the yield of a desired PCR product. Setting the annealing temperature too low may lead to amplification of nonspecific PCR products. Optimizing the annealing temperature of your PCR assay is one of the most critical parameters for reaction specificity.
#DESIGNING TOUCHDOWN PCR SOFTWARE#
Commercially available programs such as Beacon Designer software can perform both primer design and target sequence selection.
#DESIGNING TOUCHDOWN PCR FREE#
Assess primer properties (melting temperature, secondary structure, complementarity).Ī number of free online resources are available to help you with PCR assay design (see Free Internet Resources for Primer Design).Check the literature and databases for existing primers.When designing primers for a PCR assay, follow these steps:
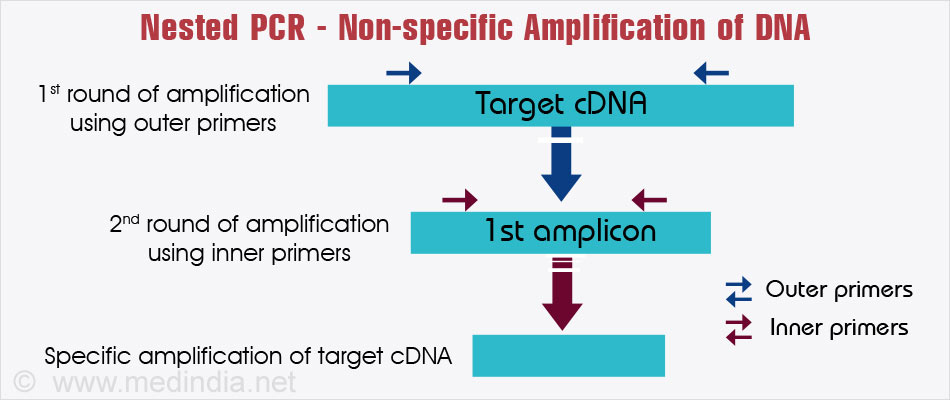
The use of PCR primers specifically designed and validated for PCR assays with your target of interest is highly recommended. Therefore, care must be taken when choosing a target sequence and designing primers. Both the primers and the target sequence can affect the efficiency, specificity, and accuracy of PCR assays. A successful PCR assay requires efficient and specific amplification of the product.


 0 kommentar(er)
0 kommentar(er)
